em-viewer¶
$ em-viewer $EM_TEST_DATA/relion_tutorial/micrographs/006.mrc --help
usage: Tool for Viewer Apps
Display the selected viewer app
positional arguments:
path
Provide a path to be visualized.
Path can be:
directory: the browser will be shown at that path.
image: an ImageView will be shown with that image.
volume: a VolumeView will be used to show the volume.
optional arguments:
-h, --help show this help message and exit
--display DISPLAY [DISPLAY ...]
Provide one or many key=value pairs with extra visualization options.
Bool values support on/off, 0/1, true/false, yes/no
Options:
axis: (bool) if false, axis will not be shown (default true)
toolbar: (bool) if false, toolbar will not be shown (default true)
histogram: (bool) if false, histogram will not shown (default true)
fit: (bool) if false, the image will not be adjusted to widget size (default true)
view: (string) Select the initial view. Options: gallery, columns, items, slices
scale: (string) Select initial scale (use % for percentage)
--visible [VISIBLE] Columns to be shown (and their order).
--render [RENDER] Columns to be rendered.
--sort [SORT] Sort command.
Viewing a Micrograph¶
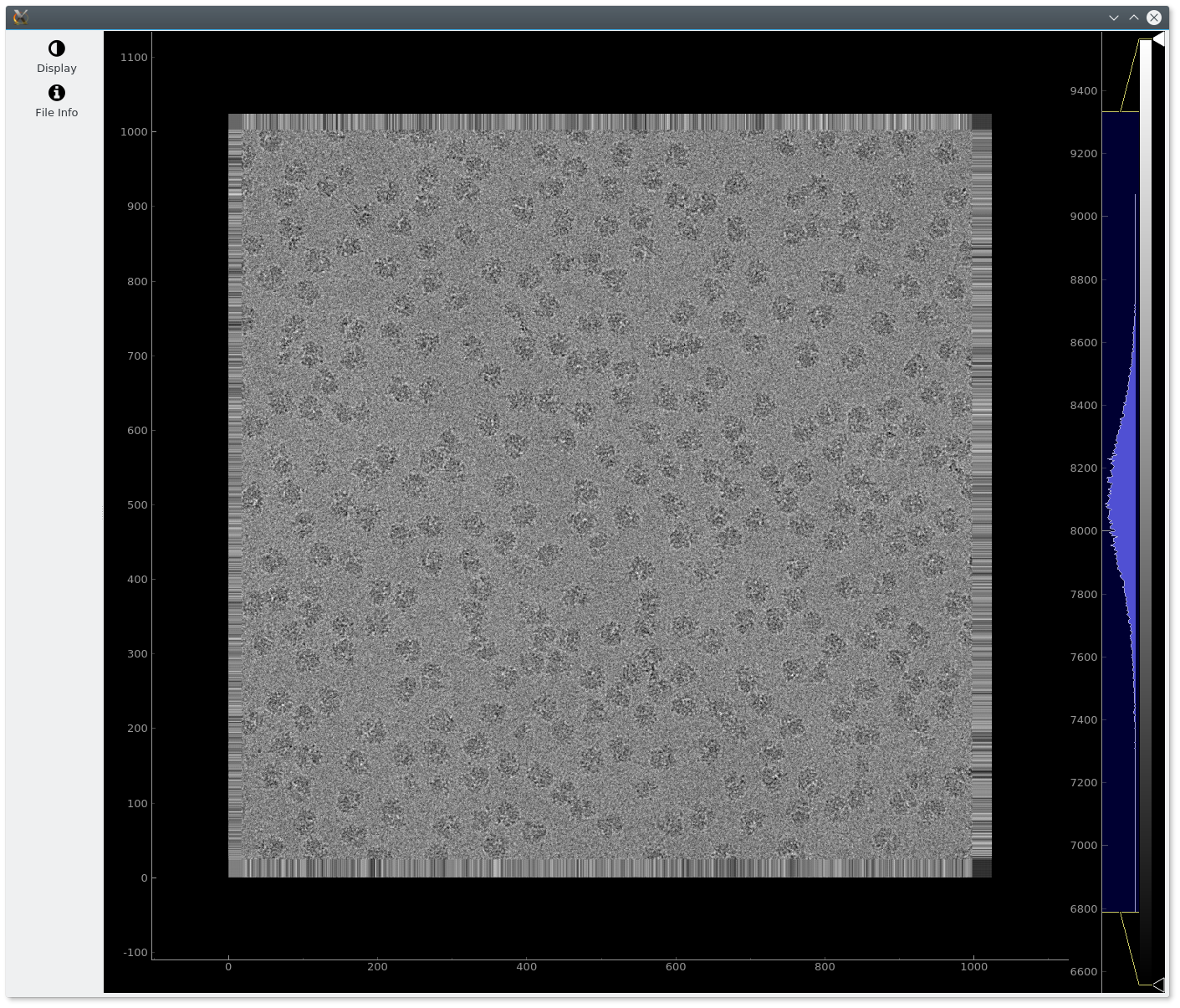
One can visualize a single image (e.g a micrograph) by providing it as the first argument.
em-viewer $EM_TEST_DATA/relion_tutorial/micrographs/006.mrc
Additionally, some extra visualization arguments can be passed via the –display option, that accepts many flags. For example, histogram=on will display by default the histogram.
em-viewer $EM_TEST_DATA/relion_tutorial/micrographs/006.mrc --display histogram=on
After this the following image should be shown:
You can also other options available to the –display option and check the result. For example:
em-viewer $EM_TEST_DATA/relion_tutorial/micrographs/006.mrc --display histogram=on axis=0 scale=0.5
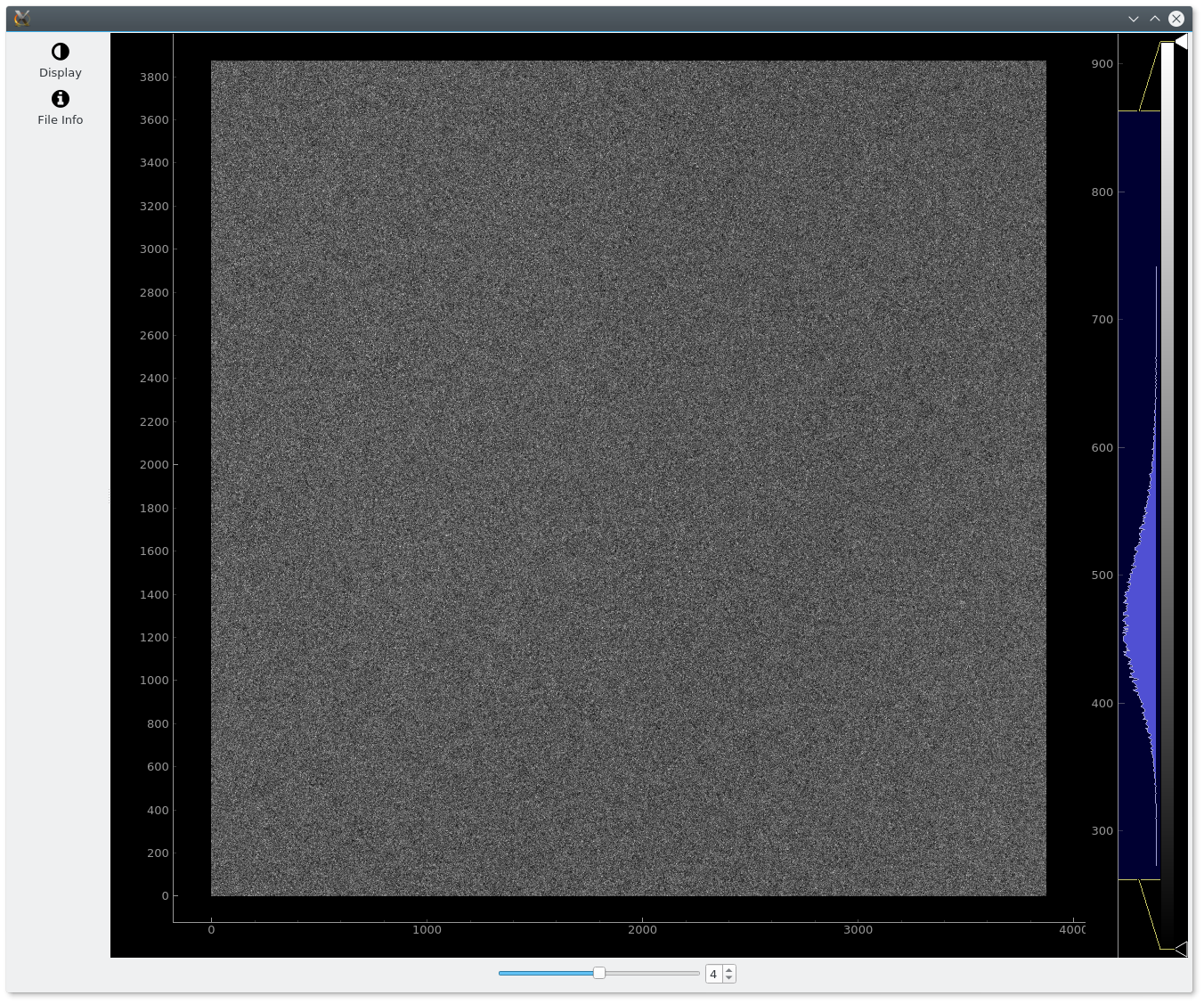
Viewing a Movie¶
Movies can be visualizing in the same way as micrographs. The display will looks very similar, but it will also contains a slider to select the frame that we want to visualize. Usually the movies are much more noisy than micrographs, but it is useful to look at it sometimes.
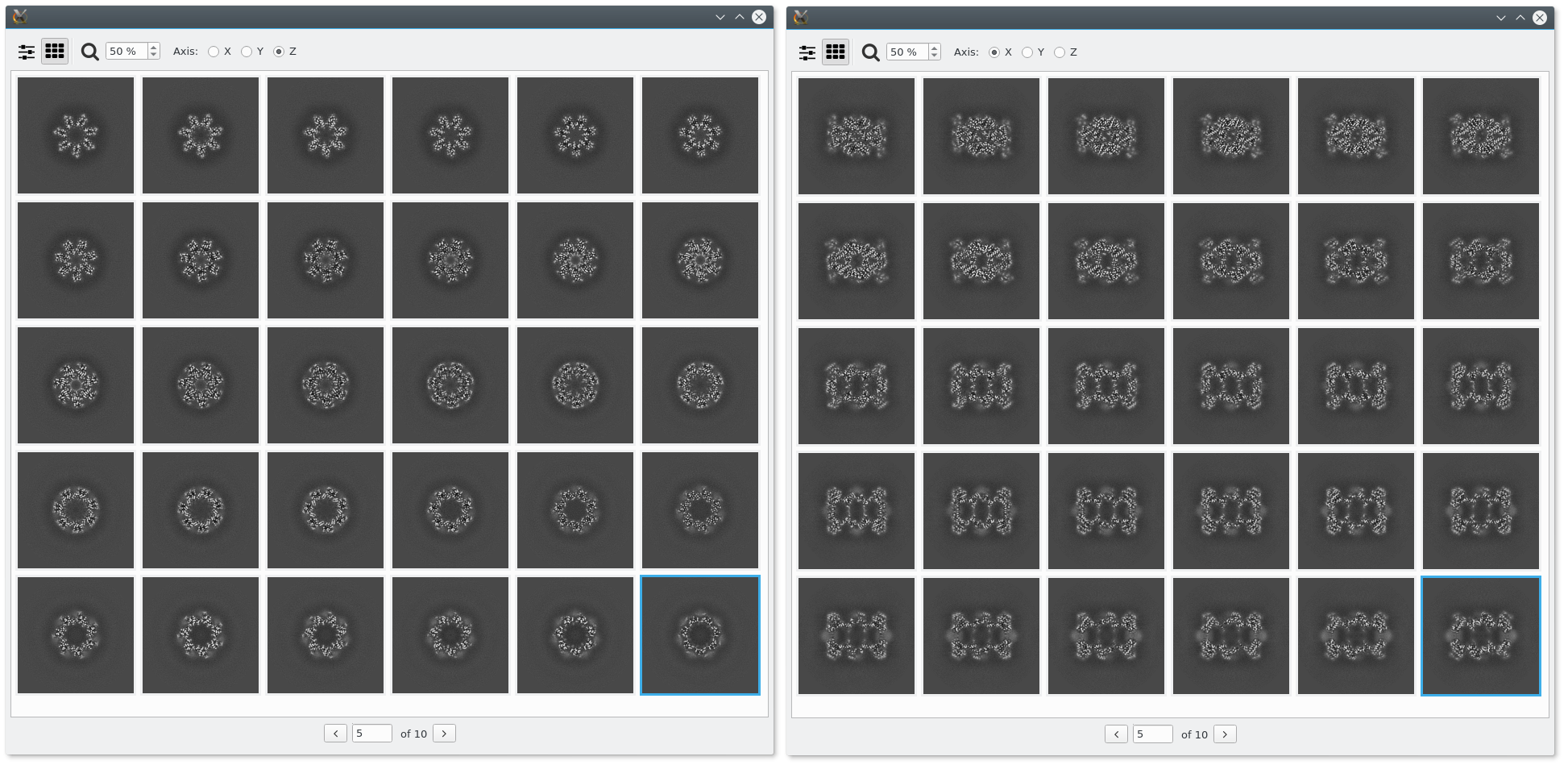
Viewing a Volume¶
Volumes can be also display with the em-viewer application. For example:
em-viewer $EM_TEST_DATA/resmap/t20s_proteasome_full.map
That will be show by default the volume with the Axis sliders view, where slices through each axis can be inspected.
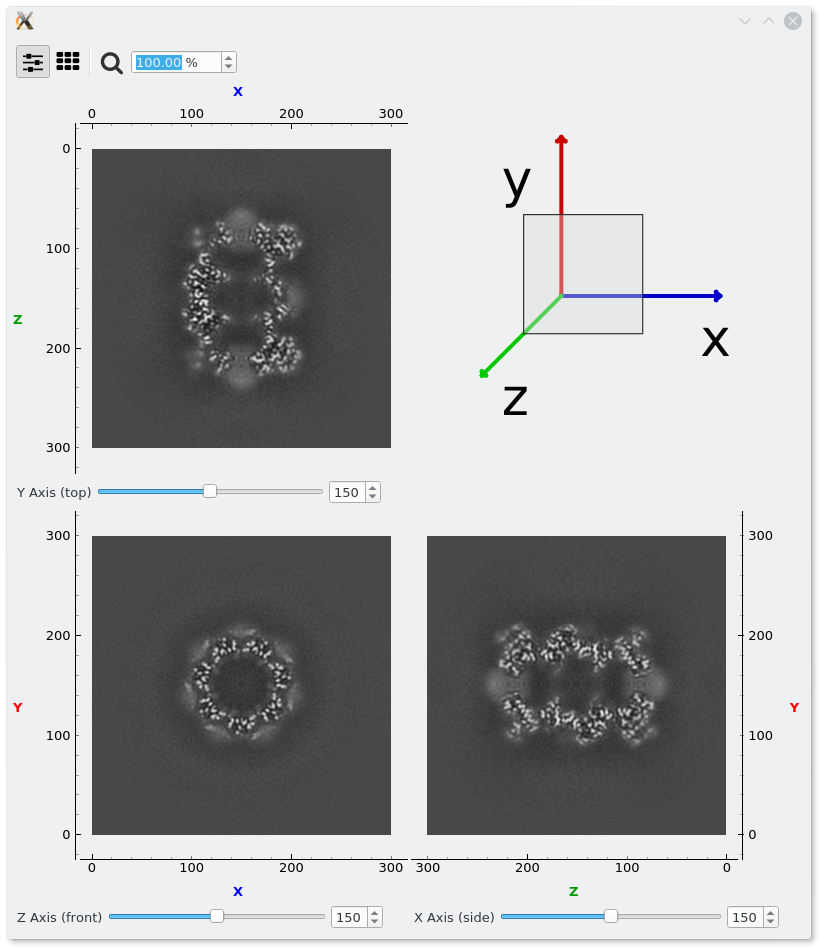
Many slices at once can be shown for one axis, and also switch between axis.